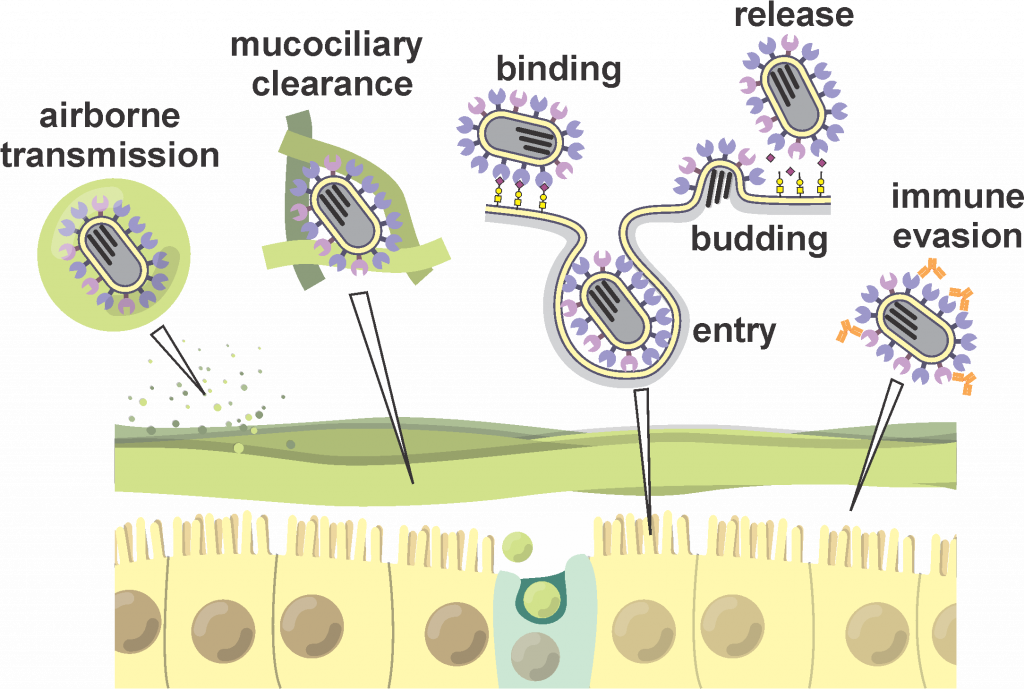
Enveloped viruses – membrane-bound pathogens including influenza, HIV, and other threats to human health – infect a wide range of hosts using a limited repertoire of molecular components. Research in the lab aims to understand how different viruses accomplish this, so that we can identify new therapeutic targets and re-engineer virus assembly to achieve new biological and therapeutic functions.
We are pursuing this goal through several interconnected and interdisciplinary projects, drawing on engineering, biology, and physics.
Reverse engineering virus assembly
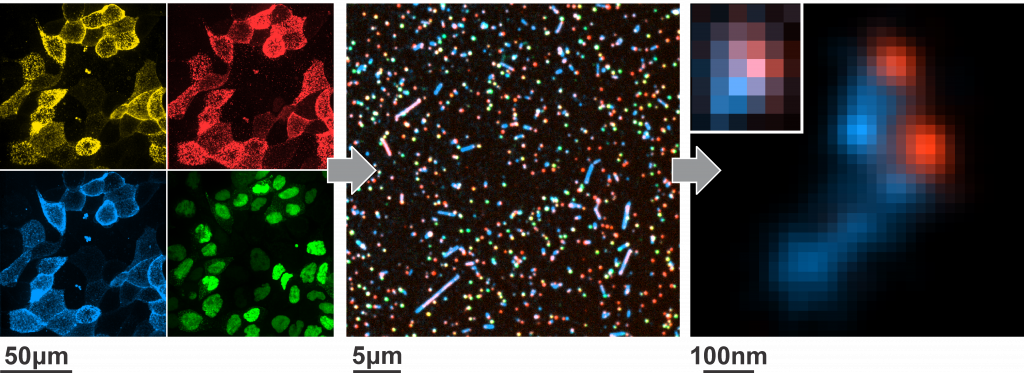
Infectiousness is a complex characteristic, arising not only from the molecules a virus contains, but also from how these molecules are organized in space and time. We use light microscopy at length scales ranging from microns to nanometers to watch viruses assemble and to non-disruptively study their organization. This information helps us to establish “design rules” that we can use to modify virus form and function.
Uncovering biophysical aspects of host-pathogen interactions
Engineering more versatile infectious disease models
We are developing model systems to monitor the progression of infection with high spatial and temporal resolution while still preserving important physiological features of the host tissues that pathogens infect.
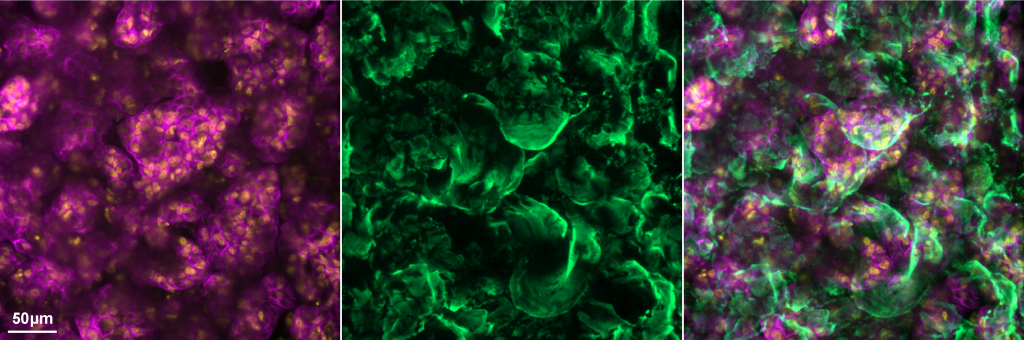 Mucus (green) secreted by differentiated airway epithelial cells (magenta & yellow) serves as a protective barrier against infection that viruses have evolved mechanisms of penetrating.
Mucus (green) secreted by differentiated airway epithelial cells (magenta & yellow) serves as a protective barrier against infection that viruses have evolved mechanisms of penetrating.